-Search query
-Search result
Showing 1 - 50 of 68 items for (author: wang & xs)

EMDB-35240: 
Cryo-EM structure of TIR-APAZ/Ago-gRNA-DNA complex
Method: single particle / : Zhang H, Deng ZQ, Yu GM, Li XZ, Wang XS

EMDB-35241: 
Cryo-EM structure of TIR-APAZ/Ago-gRNA complex
Method: single particle / : Zhang H, Li Z, Yu GM, Li XZ, Wang XS

EMDB-35592: 
Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex
Method: single particle / : Zhang H, Li Z, Yu GM, Li XZ, Wang XS
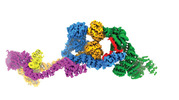
EMDB-29073: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J
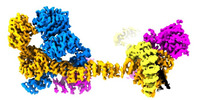
EMDB-29078: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J

PDB-8fgw: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J

PDB-8fh3: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J

EMDB-34034: 
SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up)
Method: single particle / : Wang L

EMDB-34035: 
SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down)
Method: single particle / : Wang L

EMDB-34036: 
pH 5.5 SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up)
Method: single particle / : Wang L

EMDB-34037: 
SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down)
Method: single particle / : Wang L

EMDB-34038: 
SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state1)
Method: single particle / : Wang L

EMDB-34039: 
SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state2)
Method: single particle / : Wang L

EMDB-34040: 
SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state3)
Method: single particle / : Wang L

EMDB-34041: 
SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (interface)
Method: single particle / : Wang L

EMDB-34042: 
SARS-CoV-2 BA.2.75 S Trimer in complex with XG2v024
Method: single particle / : Wang L

EMDB-34043: 
SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state1)
Method: single particle / : Wang L

EMDB-34044: 
SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state2)
Method: single particle / : Wang L

EMDB-32728: 
Local CryoEM structure of the SARS-CoV-2 S6P(B.1.1.529) in complex with BD55-4637 Fab
Method: single particle / : Du S, Xiao JY

EMDB-32737: 
Local structure of BD55-3546 Fab and SARS-COV2 Delta RBD complex
Method: single particle / : Zhang ZZ, Xiao JJ

EMDB-33552: 
Local structure of BD55-5514 and BD55-5840 Fab and Omicron BA.1 RBD complex
Method: single particle / : Zhang Z, Xiao J
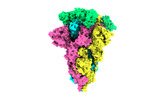
EMDB-33019: 
SARS-CoV-2 BA.2 variant spike protein in complex with Fab BD55-5840
Method: single particle / : Wang X, Wang L
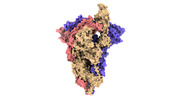
EMDB-33210: 
SARS-CoV-2 Omicron BA.2 variant spike (state 1)
Method: single particle / : Wang X, Wang L

EMDB-33211: 
SARS-CoV-2 Omicron BA.2 variant spike (state 2)
Method: single particle / : Wang X, Wang L
EMDB-33212: 
SARS-CoV-2 Omicron BA.3 variant spike
Method: single particle / : Wang X, Wang L
EMDB-33213: 
SARS-CoV-2 Omicron BA.3 variant spike (local)
Method: single particle / : Wang X, Wang L
EMDB-33323: 
SARS-CoV-2 Omicron BA.4 variant spike
Method: single particle / : Wang X, Wang L
EMDB-33324: 
SARS-CoV-2 Omicron BA.2.13 variant spike
Method: single particle / : Wang X, Wang L
EMDB-33325: 
SARS-CoV-2 Omicron BA.4 variant spike
Method: single particle / : Wang X, Wang L

EMDB-32718: 
Local CryoEM structure of the SARS-CoV-2 S6P(B.1.1.529) in complex with BD55-3152 Fab
Method: single particle / : Du S, Xiao JY

EMDB-32732: 
Local structure of BD55-1239 Fab and SARS-COV2 Omicron RBD complex
Method: single particle / : Zhang ZZ, Xiao JJ

EMDB-32734: 
Local structure of BD55-3372 and delta spike
Method: single particle / : Liu PL

EMDB-32738: 
Local resolution of BD55-5840 Fab and SARS-COV2 Omicron RBD
Method: single particle / : Zhang ZZ, Xiao JJ

EMDB-32479: 
Acidic Omicron Spike Trimer
Method: single particle / : Cui Z

EMDB-32480: 
Delta Spike Trimer(3 RBD Down)
Method: single particle / : Cui Z

EMDB-32481: 
Delta Spike Trimer(1 RBD Up)
Method: single particle / : Cui Z

EMDB-32482: 
Neutral Omicron Spike Trimer in complex with ACE2
Method: single particle / : Cui Z

EMDB-32483: 
Neutral Omicron Spike Trimer in complex with ACE2.
Method: single particle / : Cui Z

EMDB-26622: 
The V1 region of bovine V-ATPase in complex with human mEAK7 (focused refinement)
Method: single particle / : Wang R, Li X

EMDB-26623: 
CryoEM structure of a mEAK7 bound human V-ATPase complex
Method: single particle / : Wang R, Li X

EMDB-32478: 
Neutral Omicron Spike Trimer
Method: single particle / : Cui Z, Wang X

EMDB-31372: 
CryoEM map of the SARS-CoV-2 S6PV2 in complex with BD-813 Fab and BD-744 Fab
Method: single particle / : Du S, Xiao J

EMDB-31374: 
Local CryoEM structure of the SARS-CoV-2 S6PV2 in complex with BD-813 Fab and BD-744 Fab
Method: single particle / : Du S, Xiao J

EMDB-31375: 
CryoEM structure of the SARS-CoV-2 S6PV2 in complex with BD-667 Fab
Method: single particle / : Liu PL

EMDB-31376: 
CryoEM structure of the SARS-CoV-2 S6PV2 in complex with BD-771 Fab and BD-821 Fab
Method: single particle / : Zhang ZY

EMDB-31377: 
Local CryoEM structure of the SARS-CoV-2 S6PV2 in complex with BD-667
Method: single particle / : Liu PL

EMDB-31378: 
Local CryoEM structure of the SARS-CoV-2 S6PV2 in complex with BD-771 Fab and BD-821 Fab
Method: single particle / : Zhang ZY

EMDB-31379: 
CryoEM map of the SARS-CoV-2 S6PV2 in complex with BD-804 Fab
Method: single particle / : Du S, Xiao J

EMDB-31380: 
Local CryoEM structure of the SARS-CoV-2 S6PV2 in complex with BD-804 Fab
Method: single particle / : Du S, Xiao J

EMDB-31390: 
CryoEM structure of the SARS-CoV-2 S6PV2 in complex with BD-812 Fab and BD-836 Fab
Method: single particle / : Liu PL
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
